1. Statistics According to the top 100 most numerous Organisms in MERITS
In this study, we provide a visualization in the form of a word cloud displaying the 100 most commonly occurring organisms sorted by frequency. By clicking on an organism, you can access more information about it.
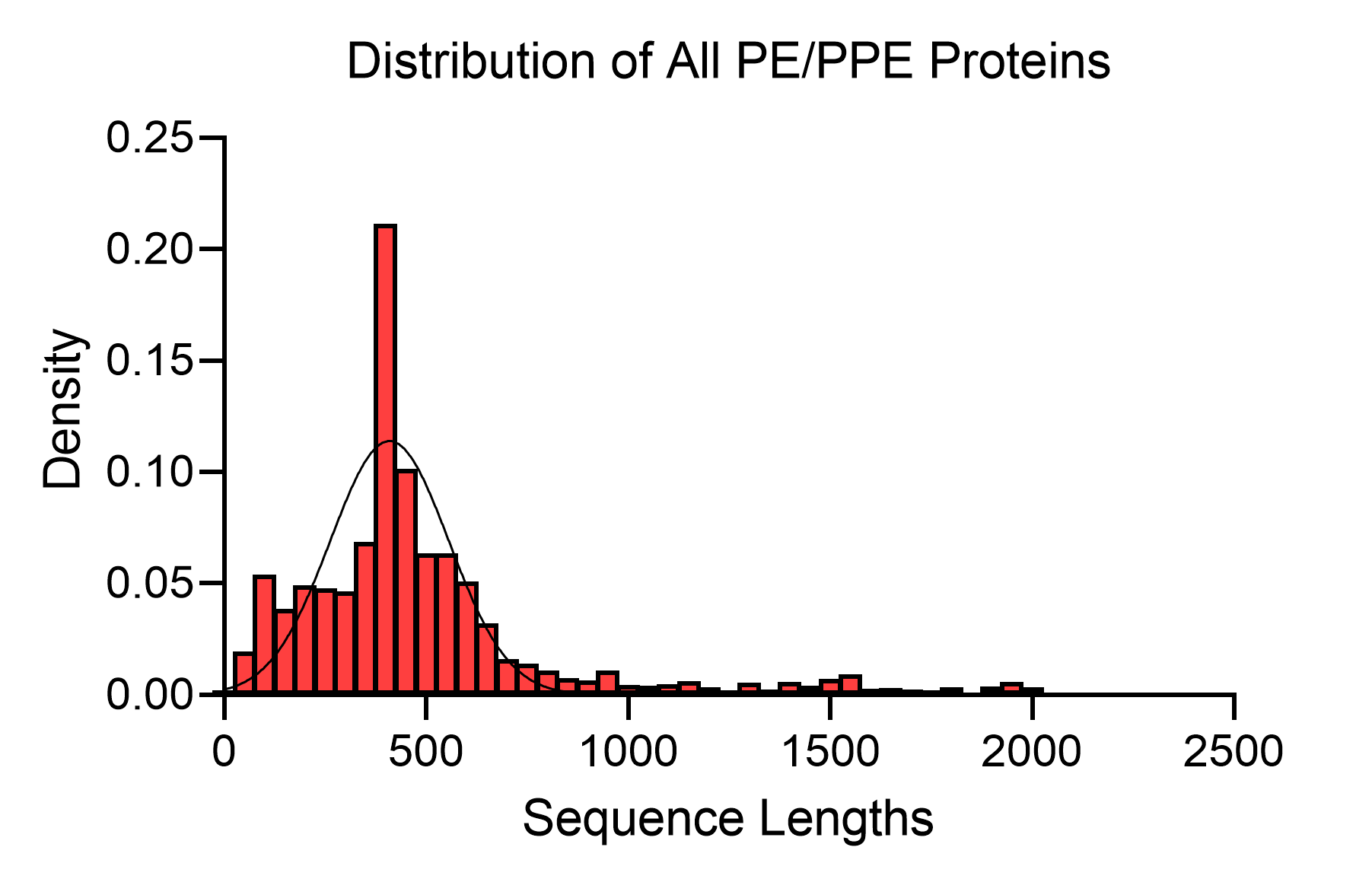
2. Distribution of all Mycobacterial PE/PPE proteins collected in MERITS based on the length of their protein sequence
Using the collected dataset, we analyzed the characteristic sequence lengths frequencies of known PE/PPE proteins. The variability in protein sequence length is a testament to the functional diversity and complexity of this protein family.
3. Frequency distributions of 20 amino acids in all accumulated Mycobacterial PE/PPE proteins.
We conducted a statistical analysis of 20 different amino acids in the database and found that glycine had the highest content, comprising 31.68% of the total.
4. Secretory protein and non-classical secretory protein prediction result in MERITS
We employed SignalP 6.0 and TMHMM 2.0 to determine if the protein sequence contained signal peptides or transmembrane domains. Subsequently, proteins that were found to contain signal peptides but not transmembrane domains are considered classically secreted. To identify non-classical protein secretion, we employed SecretomeP 2.0 computationally.
5. Phylogenetic tree of Mycobacterial PE/PPE proteins in MERITS
For each entry, the MAFFT v7.271 was used to generate multiple alignment results against Mycobacterial PE/PPE proteins in MERITS, which was visualized by the open source phylogram_d3.More than 20,000 proteins are available, but displaying them all simultaneously on the webpage would lead to suboptimal performance. Therefore, we provide newick files on the download page for users to perform their own analysis.
